-
Author
Katharina Schmolly -
PI
Simon Beaven, MD/PhD
-
Co-Author
-
Title
Reducing diagnostic delays in Acute Hepatic Porphyria (AHP) using a machine learning approach
-
Program
CTSI TL1 Summer Program
-
Other Program (if not listed above)
-
Abstract
Title: Reducing diagnostic delays in Acute Hepatic Porphyria (AHP) using a machine learning approach
Authors: Simon Beaven, MD, PhD, UCLA Principal Investigator; Kat Schmolly, UCLA MD Student Class of 2024
Affiliations: This study is a joint venture between UCLA and UCSF (Vivek Rudrapatna, MD, PhD
Bruce Wang, MD)
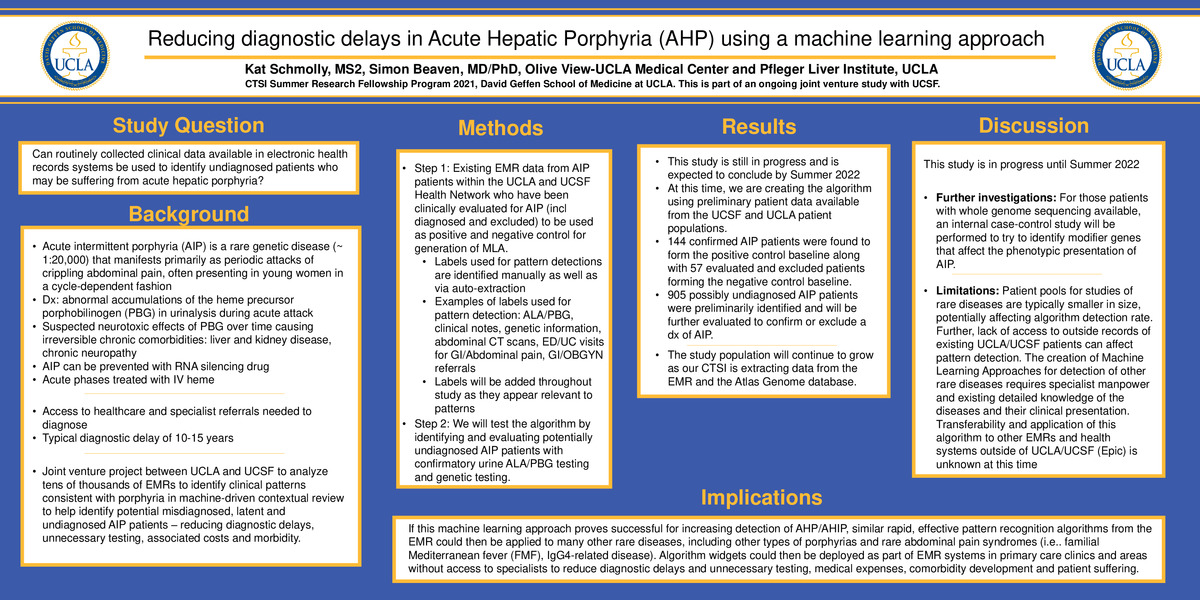
Background: Acute intermittent porphyria (AIP) is a rare genetic disease (about 1:20,000 affected) that manifests as attacks of crippling abdominal pain. Characteristic abnormal accumulations of the heme precursors delta-aminolevulinic acid (ALA) and porphobilinogen (PBG) appear to have neurotoxic effects over time, causing irreversible chronic comorbidities such as liver and kidney disease and chronic neuropathy. Because of its assumed rarity, AIP is often not considered in the differential, and thus resulting in a diagnostic delay of 10-15 years. The project is a joint venture between UCLA and UCSF to analyze routinely collected clinical data available in EMRs to attempt to reduce diagnostic delays.
Methods: Existing EMR data on AIP patients within the UCLA and UCSF Health Network who have been clinically evaluated for AIP (diagnosed and excluded) will be used to create a positive and negative control for algorithm generation. Subsequently, we will test the algorithm by identifying and evaluating potential undiagnosed AIP patients with confirmatory urine ALA/PBG testing and genetic testing.
Results: At this time, we are creating the algorithm using preliminary patient data available from the UCSF and UCLA patient populations. 144 confirmed AIP patients were found to form the positive control baseline along with 57 evaluated and excluded patients forming the negative control baseline. 905 possibly undiagnosed AIP patients were preliminarily identified to test the algorithm against.
The study population will continue to grow as our CTSI is extracting data from the EMR and the Atlas Genome database. For those patients with whole genome sequencing available, an internal case-control study will be performed to try to identify modifier genes that affect the phenotypic presentation of AIP.
Conclusion. This study is a larger joint venture project expected to conclude in Summer 2022. If successful, similar rapid, effective pattern recognition algorithms from the EMR could then be applied to many other rare diseases, including other porphyrias, familial Mediterranean fever (FMF), and IgG4-related disease.
-
PDF
-
Zoom
https://uclahs.zoom.us/j/94878982009?pwd=ZUJXOUxOS3c1bjU1NlpaZ21ZNVZIQT09